S-96 Chapter 8 Nucleotides and Nucleic Acids
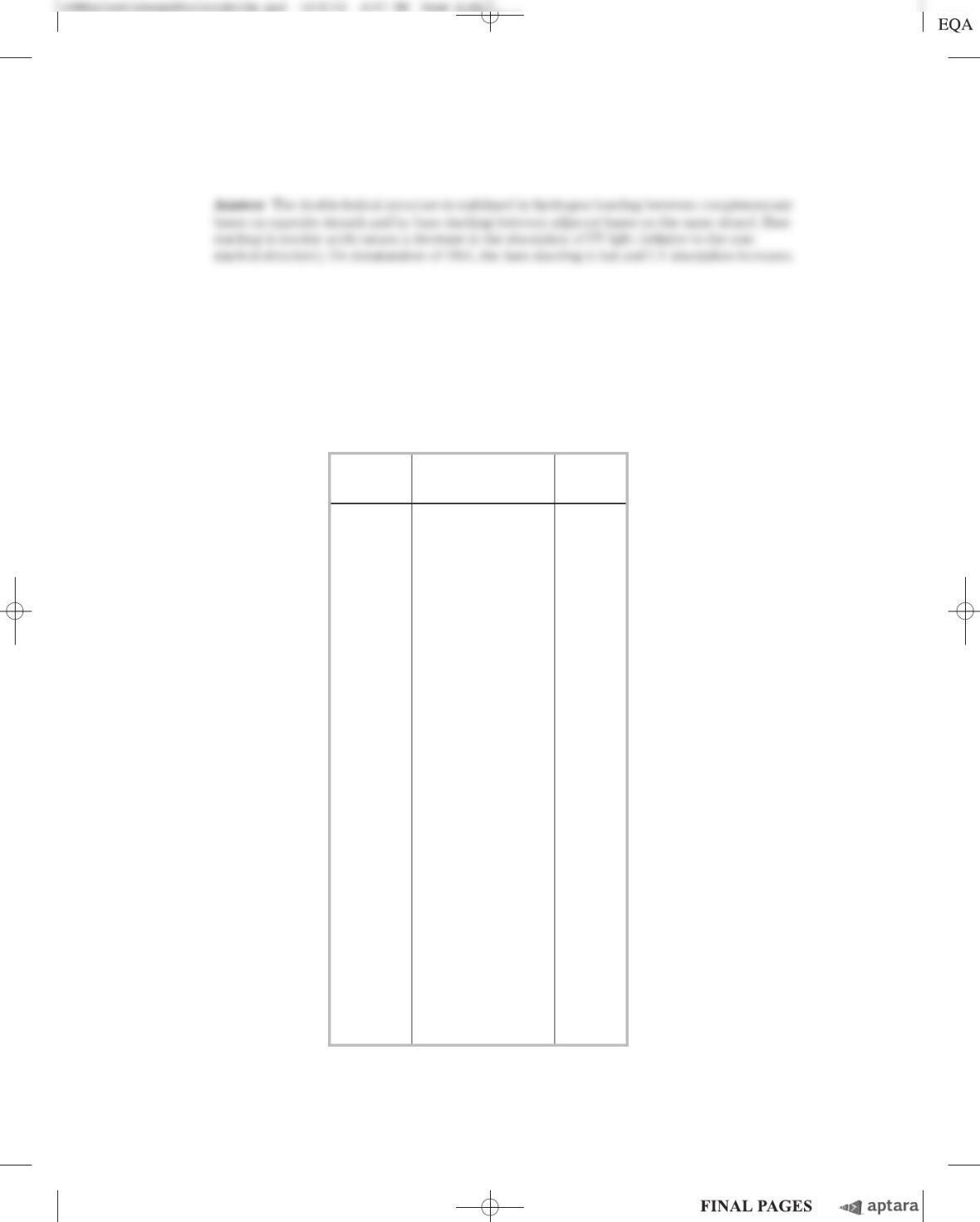
(a) Based on these data, Chargaff concluded that “no differences in composition have so far been
found in DNA from different tissues of the same species.” This corresponds to conclusion 2 in
this chapter. However, a skeptic looking at the data above might say, “They certainly look differ-
ent to me!” If you were Chargaff, how would you use the data to convince the skeptic to change
her mind?
(b) The base composition of DNA from normal and cancerous liver cells (hepatocarcinoma) was not
distinguishably different. Would you expect Chargaff’s technique to be capable of detecting a dif-
ference between the DNA of normal and cancerous cells? Explain your reasoning.
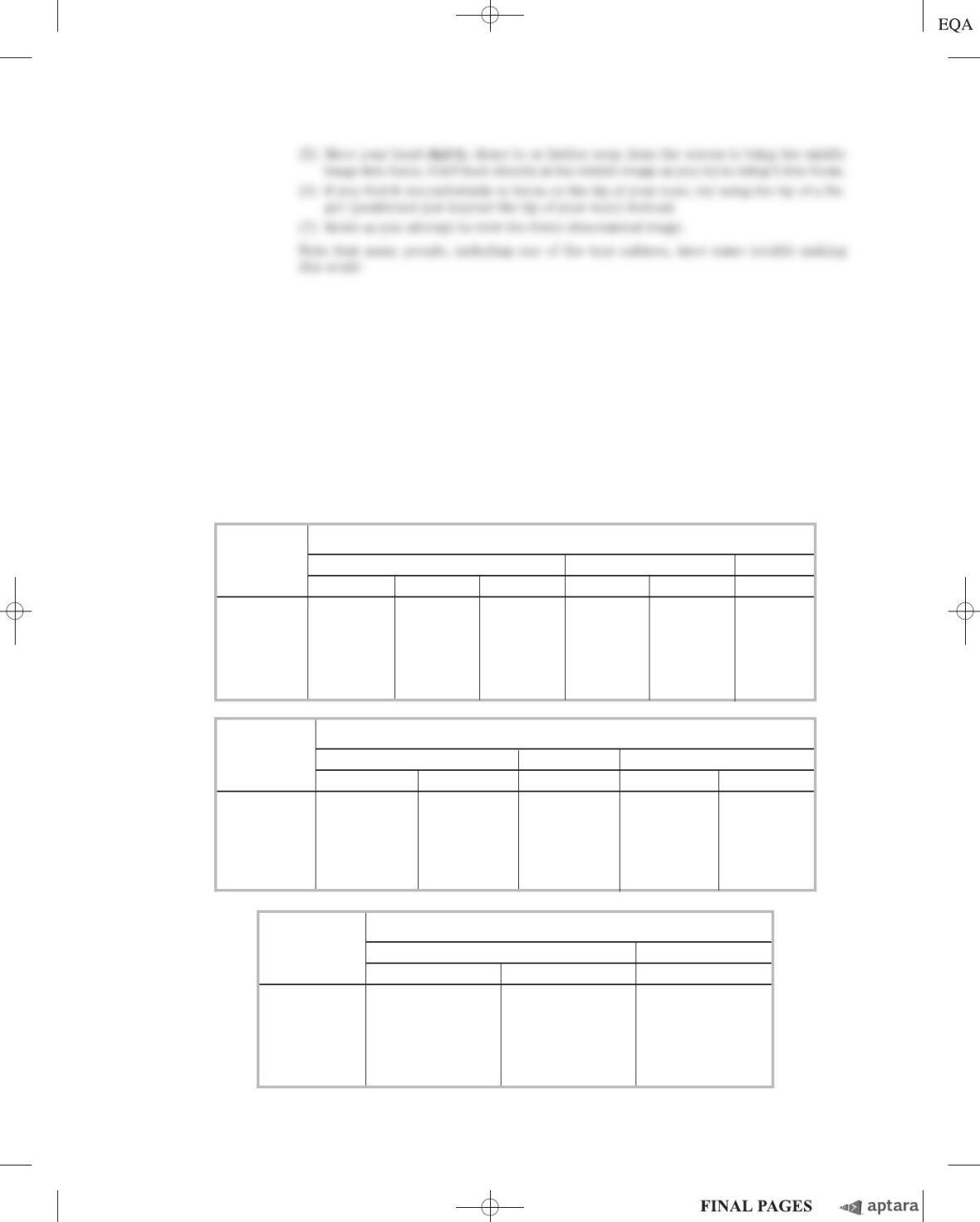
As you might expect, Chargaff’s data were not completely convincing. He went on to improve his
techniques, as described in his 1951 paper, in which he reported molar ratios of bases in DNA from a
variety of organisms:
Source A:G T:C A:T G:C Purine:pyrimidine
Ox 1.29 1.43 1.04 1.00 1.1
Human 1.56 1.75 1.00 1.00 1.0
Hen 1.45 1.29 1.06 0.91 0.99
Salmon 1.43 1.43 1.02 1.02 1.02
Wheat 1.22 1.18 1.00 0.97 0.99
Yeast 1.67 1.92 1.03 1.20 1.0
Haemophilus
influenzae
type c 1.74 1.54 1.07 0.91 1.0
E. coli
K-12 1.05 0.95 1.09 0.99 1.0
Avian tubercle
bacillus 0.4 0.4 1.09 1.08 1.1
Serratia
marcescens
0.7 0.7 0.95 0.86 0.9
Bacillus schatz
0.7 0.6 1.12 0.89 1.0
(c) According to Chargaff, as stated in conclusion 1 in this chapter, “The base composition of DNA
generally varies from one species to another.” Provide an argument, based on the data presented
so far, that supports this conclusion.
(d) According to conclusion 4, “In all cellular DNAs, regardless of the species . . . A ⫹G ⫽T ⫹C.”
Provide an argument, based on the data presented so far, that supports this conclusion.
Part of Chargaff’s intent was to disprove the “tetranucleotide hypothesis”; this was the idea that DNA
was a monotonous tetranucleotide polymer (AGCT)
n
and therefore not capable of containing sequence in-
formation. Although the data presented above show that DNA cannot be simply a tetranucleotide—if so, all
samples would have molar ratios of 0.25 for each base—it was still possible that the DNA from different or-
ganisms was a slightly more complex, but still monotonous, repeating sequence.
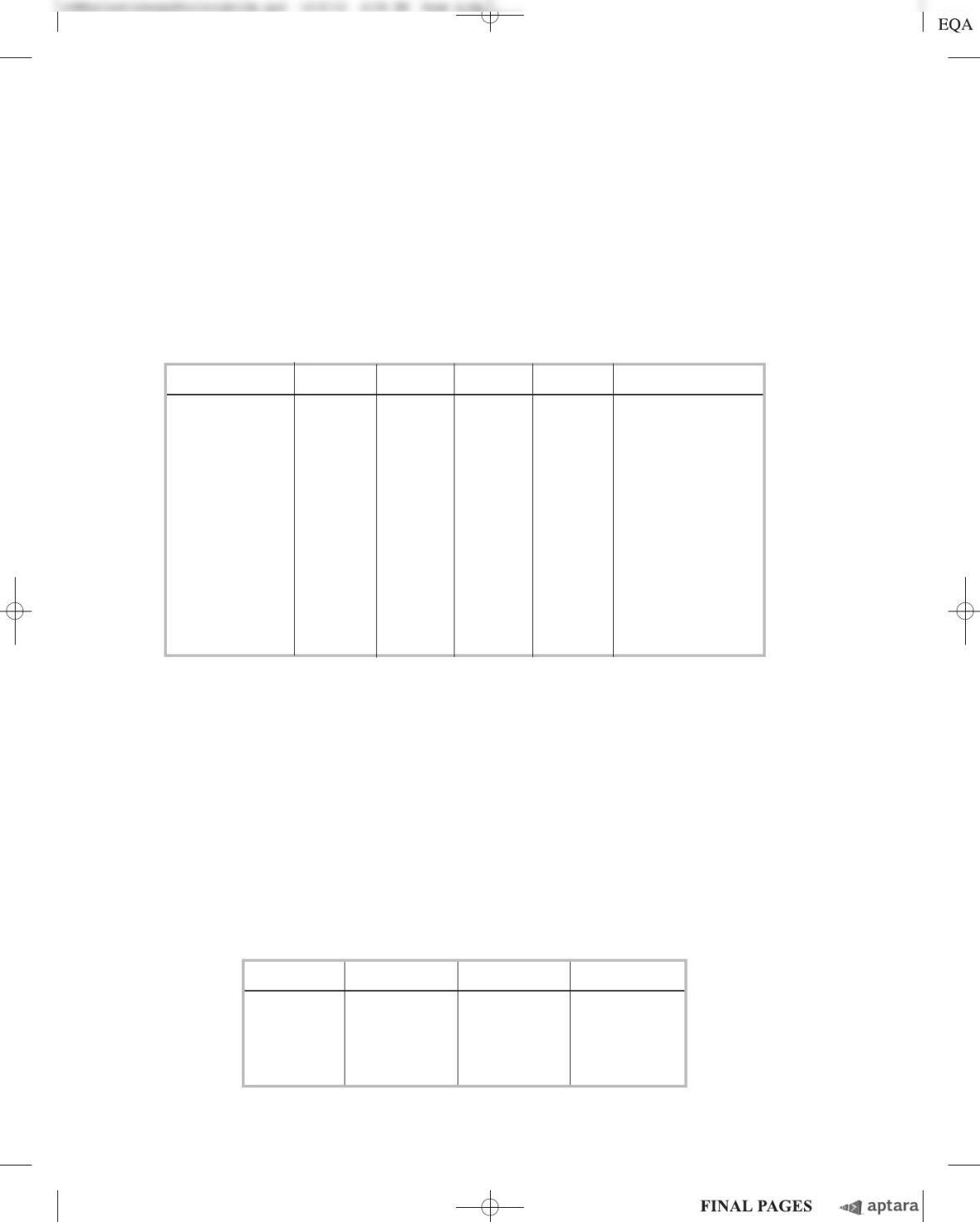
To address this issue, Chargaff took DNA from wheat germ and treated it with the enzyme de-
oxyribonuclease for different time intervals. At each time interval, some of the DNA was converted to
small fragments; the remaining, larger fragments he called the “core.” In the table below, the “19%
core” corresponds to the larger fragments left behind when 81% of the DNA was degraded; the “8%
core” corresponds to the larger fragments left after 92% degradation.
Base Intact DNA 19% Core 8% Core
Adenine 0.27 0.33 0.35
Guanine 0.22 0.20 0.20
Cytosine 0.22 0.16 0.14
Thymine 0.27 0.26 0.23
Recovery 0.98 0.95 0.92