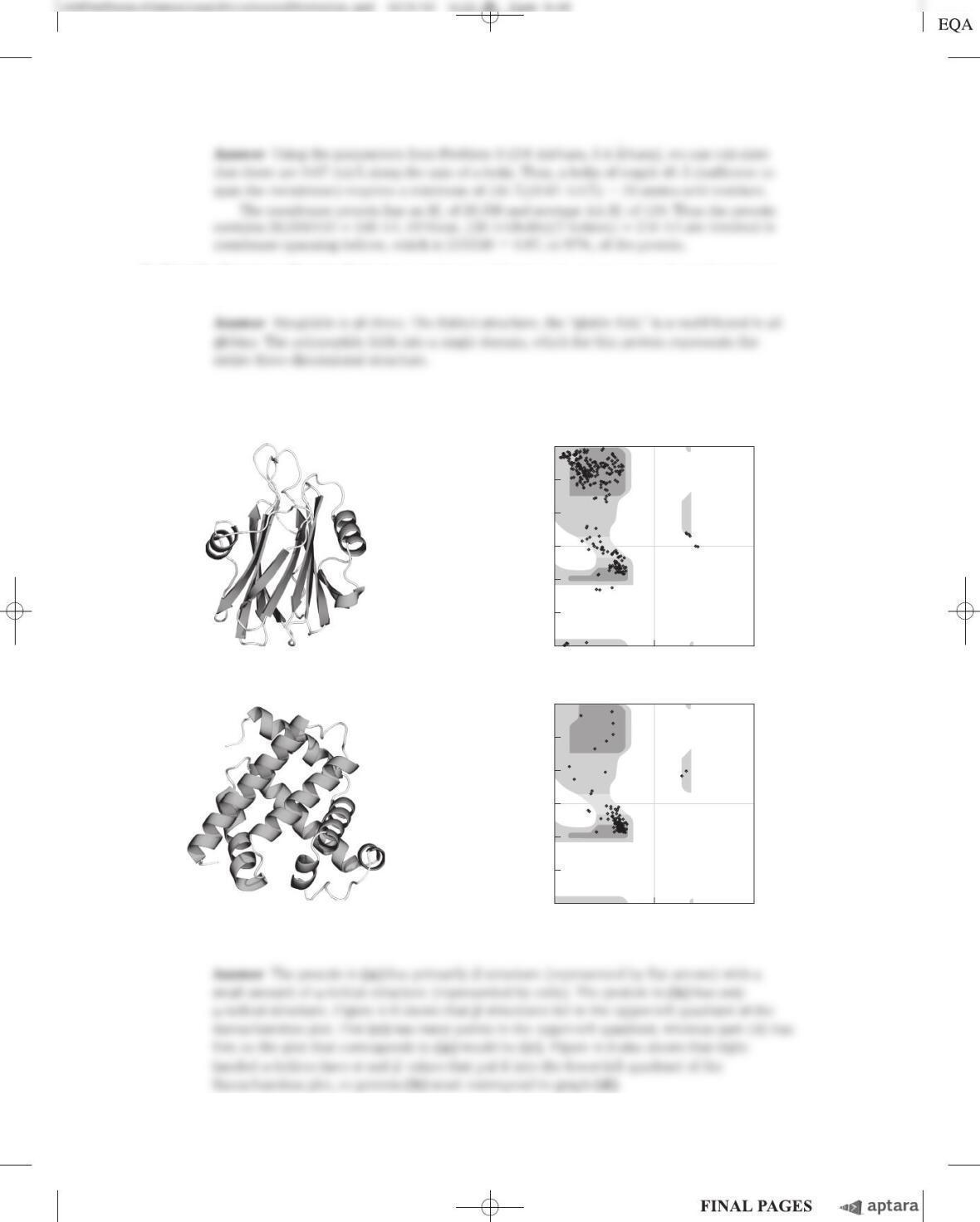
S-46 Chapter 4 The Three-Dimensional Structure of Proteins
What is the explanation for the effect of the pH changes on the conformations of poly(Glu) and
poly(Lys)? Why does the transition occur over such a narrow range of pH?
5. Disulfide Bonds Determine the Properties of Many Proteins Some natural proteins are rich in
disulfide bonds, and their mechanical properties (tensile strength, viscosity, hardness, etc.) are corre-
lated with the degree of disulfide bonding.
(a) Glutenin, a wheat protein rich in disulfide bonds, is responsible for the cohesive and elastic char-
acter of dough made from wheat flour. Similarly, the hard, tough nature of tortoise shell is due to
the extensive disulfide bonding in its a-keratin. What is the molecular basis for the correlation
between disulfide-bond content and mechanical properties of the protein?
(b) Most globular proteins are denatured and lose their activity when briefly heated to 65 C.
However, globular proteins that contain multiple disulfide bonds often must be heated longer at
higher temperatures to denature them. One such protein is bovine pancreatic trypsin inhibitor
(BPTI), which has 58 amino acid residues in a single chain and contains three disulfide bonds.
On cooling a solution of denatured BPTI, the activity of the protein is restored. What is the
molecular basis for this property?
Answer
(b) As the temperature is raised, the increased thermal motion of the polypeptide chains
6. Dihedral Angles A series of torsion angles, and , that might be taken up by the peptide backbone
is shown below. Which of these closely correspond to and for an idealized collagen triple helix?
Refer to Figure 4
–
9 as a guide.
(a) (b) (c) (d) (e) (f)