Chapter 3 Amino Acids, Peptides, and Proteins S-37
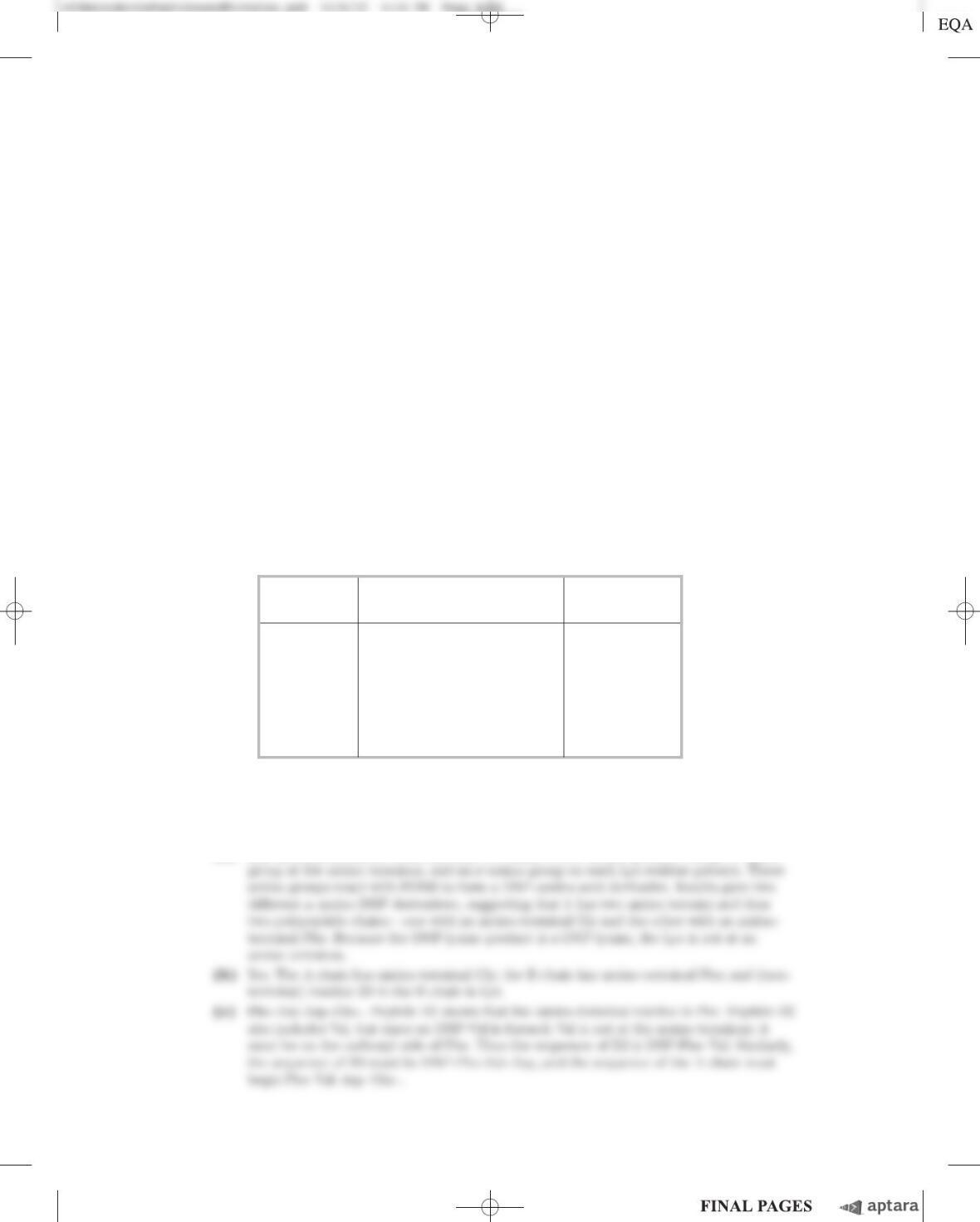
(a) From the information given in the table, calculate the specific activity of the enzyme after each
purification procedure.
(b) Which of the purification procedures used for this enzyme is most effective (i.e., gives the greatest
relative increase in purity)?
(c) Which of the purification procedures is least effective?
(d) Is there any indication based on the results shown in the table that the enzyme after step 6 is
now pure? What else could be done to estimate the purity of the enzyme preparation?
Answer
16. Dialysis A purified protein is in a Hepes (N-(2-hydroxyethyl)piperazine-N⬘-(2-ethanesulfonic acid)) buffer
at pH 7 with 500 m
M
NaCl. A sample (1 mL) of the protein solution is placed in a tube made of dialysis mem-
brane and dialyzed against 1 L of the same Hepes buffer with 0 m
M
NaCl. Small molecules and ions (such as
Na
⫹
, Cl
⫺
, and Hepes) can diffuse across the dialysis membrane, but the protein cannot.
(a) Once the dialysis has come to equilibrium, what is the concentration of NaCl in the protein sam-
ple? Assume no volume changes occur in the sample during the dialysis.
(b) If the original 1 mL sample were dialyzed twice, successively, against 100 mL of the same Hepes
buffer with 0 m
M
NaCl, what would be the final NaCl concentration in the sample?
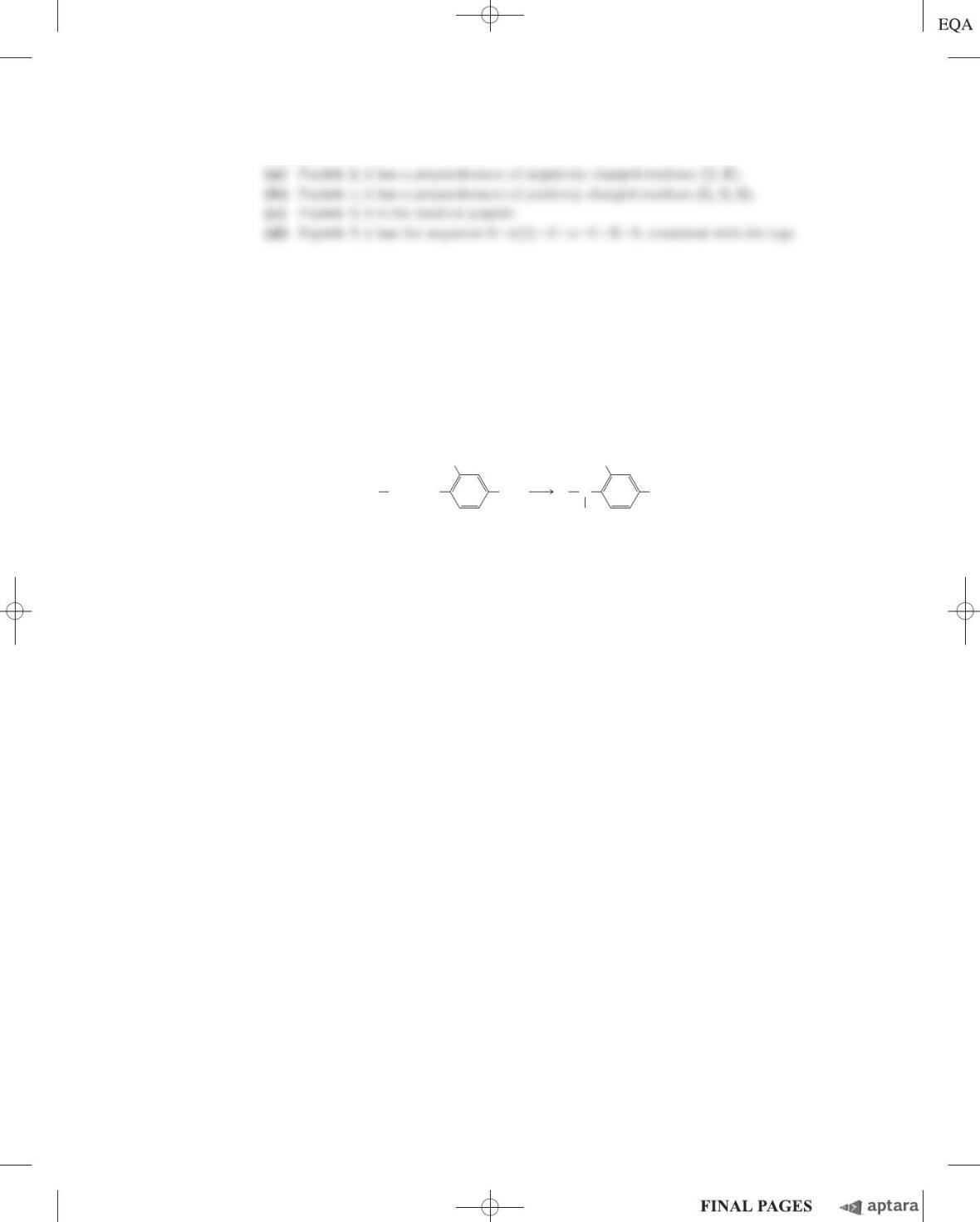
17. Peptide Purification At pH 7.0, in what order would the following three peptides be eluted from a col-
umn filled with a cation-exchange polymer? Their amino acid compositions are:
Peptide A: Ala 10%, Glu 5%, Ser 5%, Leu 10%, Arg 10%, His 5%, Ile 10%, Phe 5%, Tyr 5%, Lys 10%,
Gly 10%, Pro 5%, and Trp 10%.
Peptide B: Ala 5%, Val 5%, Gly 10%, Asp 5%, Leu 5%, Arg 5%, Ile 5%, Phe 5%, Tyr 5%, Lys 5%,
Trp 5%, Ser 5%, Thr 5%, Glu 5%, Asn 5%, Pro 10%, Met 5%, and Cys 5%.
Peptide C: Ala 10%, Glu 10%, Gly 5%, Leu 5%, Asp 10%, Arg 5%, Met 5%, Cys 5%, Tyr 5%, Phe 5%,
His 5%, Val 5%, Pro 5%, Thr 5%, Ser 5%, Asn 5%, and Gln 5%.