Chapter 28 Regulation of Gene Expression S-317
Data Analysis Problem
15. Engineering a Genetic Toggle Switch in Escherichia coli Gene regulation is often described as
an “on or off” phenomenon—a gene is either fully expressed or not expressed at all. In fact, repression
and activation of a gene involve ligand-binding reactions, so genes can show intermediate levels of
expression when intermediate levels of regulatory molecules are present. For example, for the E. coli
lac operon, consider the binding equilibrium of the Lac repressor, operator DNA, and inducer (see
Fig. 28–8). Although this is a complex, cooperative process, it can be approximately modeled by the
following reaction (R is repressor; IPTG is the inducer isopropyl--
D
-thiogalactoside):
R IPTG u99999999v R IPTG
Free repressor, R, binds to the operator and prevents transcription of the lac operon; the R IPTG
complex does not bind to the operator and thus transcription of the lac operon can proceed.
(a) Using Equation 5–8, we can calculate the relative expression level of the proteins of the lac
operon as a function of [IPTG]. Use this calculation to determine over what range of [IPTG] the
expression level would vary from 10% to 90%.
(b) Describe qualitatively the level of lac operon proteins present in an E. coli cell before, during,
and after induction with IPTG. You need not give the amounts at exact times—just indicate the
general trends.
Gardner, Cantor, and Collins (2000) set out to make a “genetic toggle switch”—a gene-regulatory
system with two key characteristics of a light switch. (A) It has only two states: it is either fully on
or fully off; it is not a dimmer switch. In biochemical terms, the target gene or gene system (operon)
is either fully expressed or not expressed at all; it cannot be expressed at an intermediate level. (B)
Both states are stable: although you must use a finger to flip the light switch from one state to the
other, once you have flipped it and removed your finger, the switch stays in that state. In biochemical
terms, exposure to an inducer or some other signal changes the expression state of the gene or
operon, and it remains in that state once the signal is removed.
(c) Explain how the lac operon lacks both characteristics A and B.
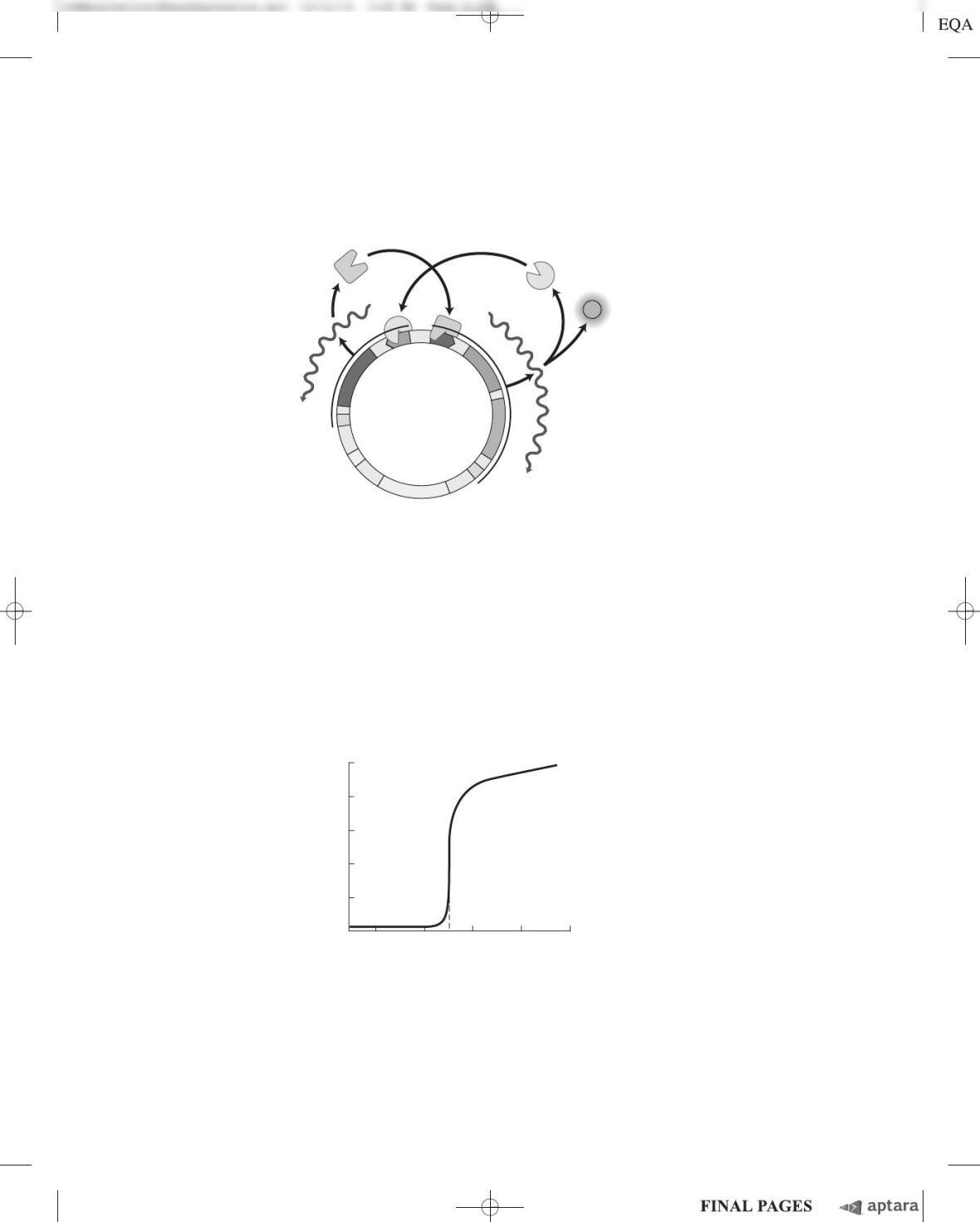
To make their “toggle switch,” Gardner and coworkers constructed a plasmid from the
following components:
OP
lac
The operator-promoter region of the E. coli lac operon
OP
The operator-promoter region of phage
lacI The gene encoding the lac repressor protein, LacI. In the absence of IPTG, this protein
strongly represses OP
lac
; in the presence of IPTG, it allows full expression from OP
lac
.
rep
ts
The gene encoding a temperature-sensitive mutant repressor protein, rep
ts
. At 37 C this
protein strongly represses OP
; at 42 C it allows full expression from OP
.
GFP The gene for green fluorescent protein (GFP), a highly fluorescent reporter protein (see
Fig. 9–16)
T Transcription terminator
K
d
10
4
M