S-292 Chapter 25 DNA Metabolism
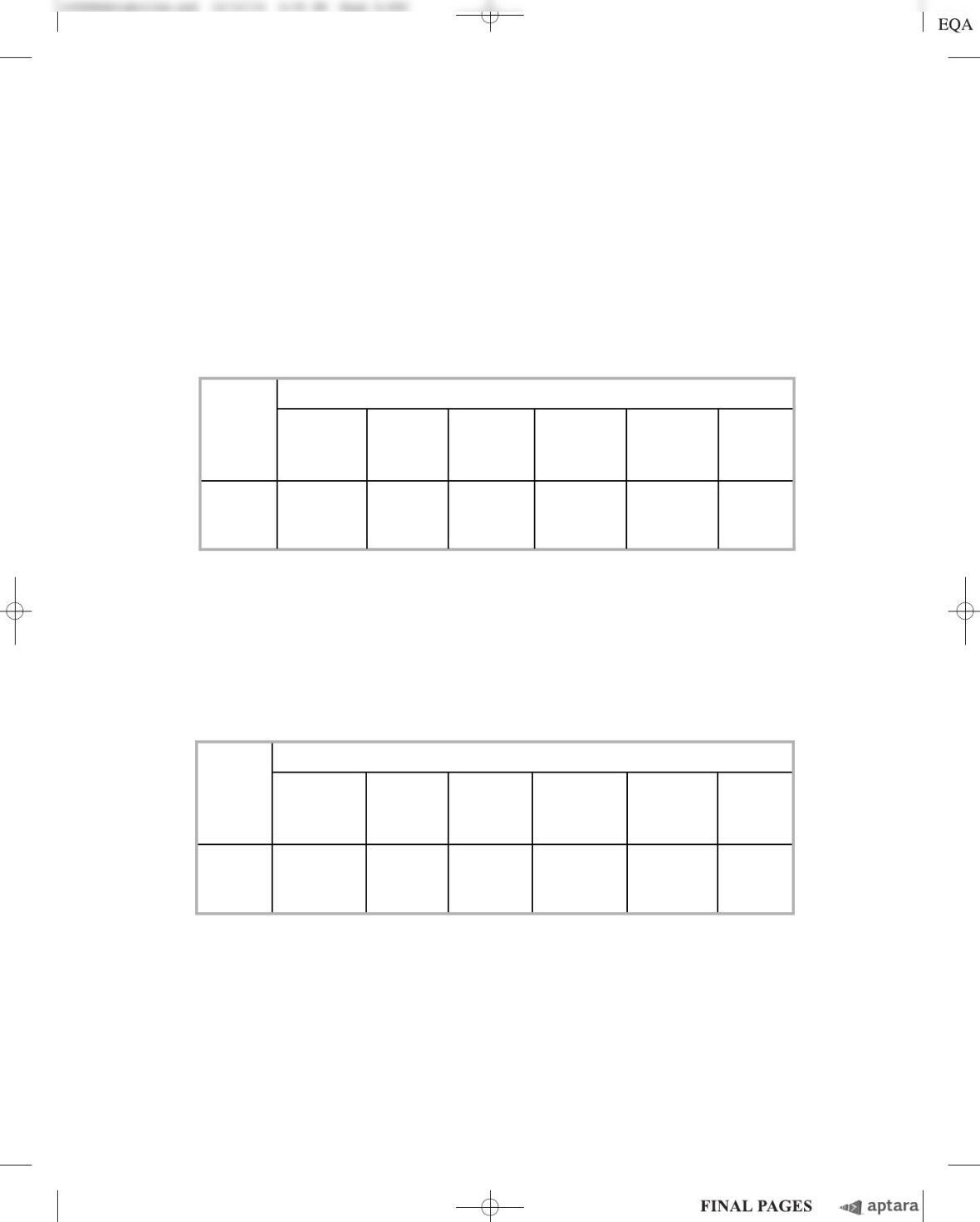
Number of Lac
ⴙ
cells (average ⴞSD)
CC101 CC102 CC103 CC104 CC105 CC106
(AUT(GmC(GmC (GmC(AUT(AUT
R7000 to to to to to to
(g/mL) CmG) AUT) CmG) TUA) TUA) GmC)
02 2 10 93 34 26 10.5 1
17 6 21 98 3 23 15 13 11 1
54 3 15 7 22 2 68 25 67 14 1 1
Number of Lac
ⴙ
cells (average ⴞSD)
CC101 CC102 CC103 CC104 CC105 CC106
(AUT (GmC (GmC (GmC (AUT (AUT
R7000 to to to to to to
(g/mL) CmG) AUT) CmG) TUA) TUA) GmC)
06 3 11 92 15 32 11 1
0.075 24 19 34 38 4 82 23 40 14 4 2
0.15 24 4 26 29 5 180 71 130 50 3 2
Quillardet and colleagues then examined the particular DNA sequence changes caused by
R7000 in the uvr
and uvr
bacteria. For this, they used six different strains of E. coli, each with a
different point mutation in the lacZ gene, which encodes -galactosidase (this enzyme catalyzes the
same reaction as lactase; see Fig. 14–11). Cells with any of these mutations have a nonfunctional
-galactosidase and are unable to metabolize lactose (i.e., a Lac
phenotype). Each type of point
mutation required a specific reverse mutation to restore lacZ gene function and Lac
phenotype. By
plating cells on a medium containing lactose as the sole carbon source, it was possible to select for
these reverse-mutated, Lac
cells. And by counting the number of Lac
cells following mutagenesis
of a particular strain the researchers could measure the frequency of each type of mutation.
First, they looked at the mutation spectrum in uvr
cells. The following table shows the results
for the six strains, CC101 through CC106 (with the point mutation required to produce Lac
cells in-
dicated in parentheses).
(e) Which types of mutation show significant increases above the background rate due to treatment
with R7000? Provide a plausible explanation for why some have higher frequencies than others.
(f) Can all of the mutations you listed in (e) be explained as resulting from covalent attachment of
R7000 to a GmC base pair? Explain your reasoning.
(g) Figure 25–27b shows how methylation of guanine residues can lead to a GmC to APT mutation. Using a
similar pathway, show how a G–R7000 adduct could lead to the GmC to APT or TPA mutations shown
above. Which base pairs with the GOR7000 adduct?
The results for the uvr
bacteria are shown in the table below.
(h) Do these results show that all mutation types are repaired with equal fidelity? Provide a plausible expla-
nation for your answer.