5. Studying genes and their products
Certain restriction enzymes produce cohesive (sticky) ends. This means that they:
A) cut both DNA strands at the same base pair.
B) cut in regions of high GC content, leaving ends that can form more hydrogen bonds than ends of
high AT content.
C) make a staggered double-strand cut, leaving ends with a few nucleotides of single-stranded DNA
protruding.
D) make ends that can anneal to cohesive ends generated by any other restriction enzyme.
E) stick tightly to the ends of the DNA they have cut.
6. Studying genes and their products
In the laboratory, recombinant plasmids are commonly introduced into bacterial cells by:
A) electrophoresis—a gentle low-voltage gradient draws the DNA into the cell.
B) infection with a bacteriophage that carries the plasmid.
C) microinjection.
D) mixing plasmids with an extract of broken cells.
E) transformation—heat shock of the cells incubated with plasmid DNA in the presence of CaCl2.
7. Studying genes and their products
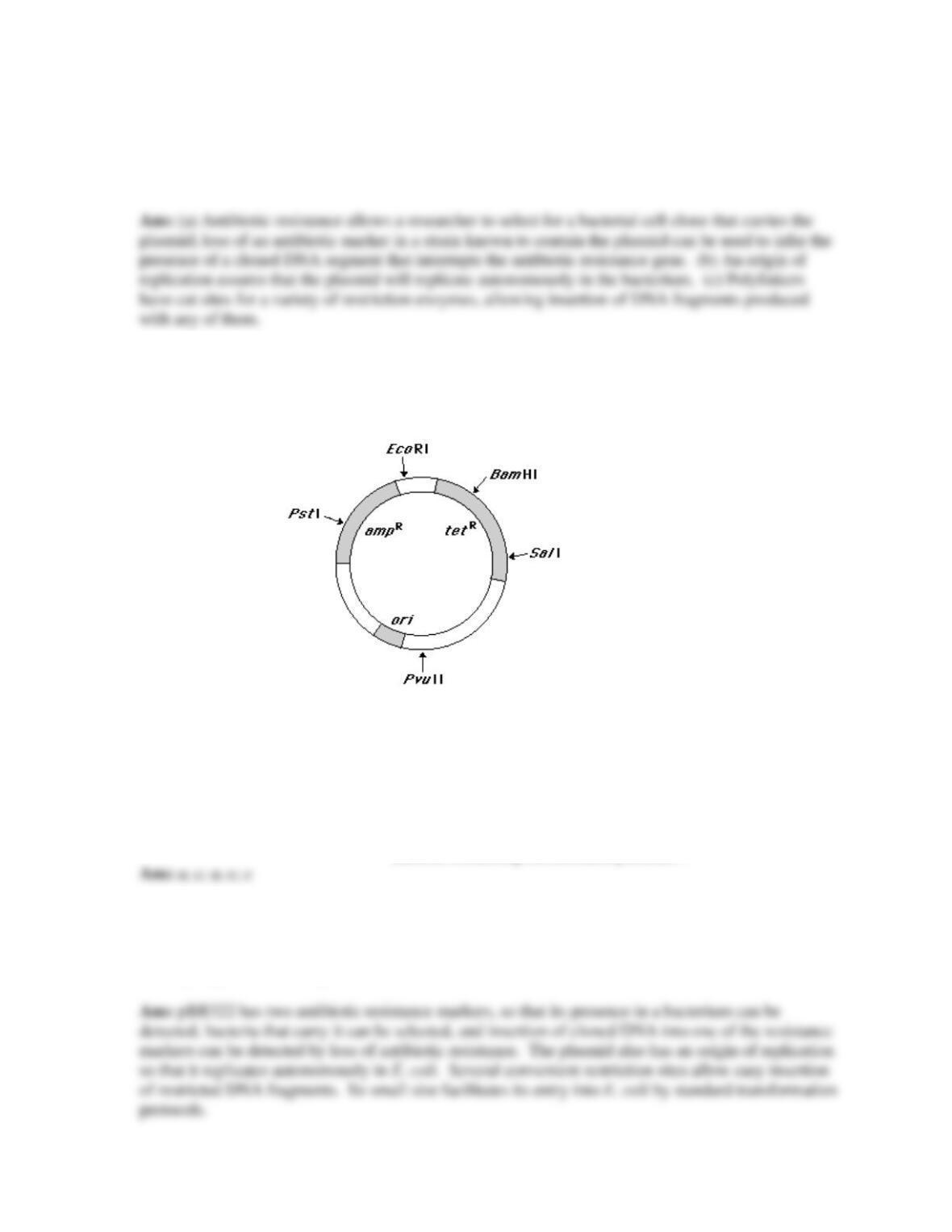
The E. coli recombinant plasmid pBR322 has been widely utilized in genetic engineering
experiments. pBR322 has all of the following features except:
A) a number of conveniently located recognition sites for restriction enzymes.
B) a number of palindromic sequences near the EcoRI site, which permit the plasmid to assume a
conformation that protects newly inserted DNA from nuclease degradation.
C) a replication origin, which permits it to replicate autonomously.
D) resistance to two different antibiotics, which permits rapid screening for recombinant plasmids
containing foreign DNA.
E) small overall size, which facilitates entry of the plasmid into host cells.
8. Studying genes and their products
Which of the following statements regarding plasmid-cloning vectors is correct?
A) Circular plasmids do not require an origin of replication to be propagated in E. coli.
B) Foreign DNA fragments up to 45,000 base pairs can be cloned in a typical plasmid.
C) Plasmids do not need to contain genes that confer resistance to antibiotics.
D) Plasmid vectors must carry promoters for inserted gene fragments.
E) The copy number of plasmids may vary from a few to several hundred.