18
64) Some restriction enzymes do not leave sticky ends when cutting DNA molecules; rather,
they cut a restriction site down the middle and leave "blunt-ended" DNA molecules, which do
not have any single-stranded nucleotide extensions. Which of the following is not a likely
outcome when trying to insert a gene into a plasmid when have both been cut with the same
"blunt-ended" restriction enzyme?
A) The gene might be inserted into the plasmid by forming hydrogen bonds.
B) The gene might be inserted into the plasmid multiple times in a row.
C) The gene might insert into the plasmid in the proper (forward) orientation.
D) The gene might insert into the plasmid in the wrong (backward) orientation.
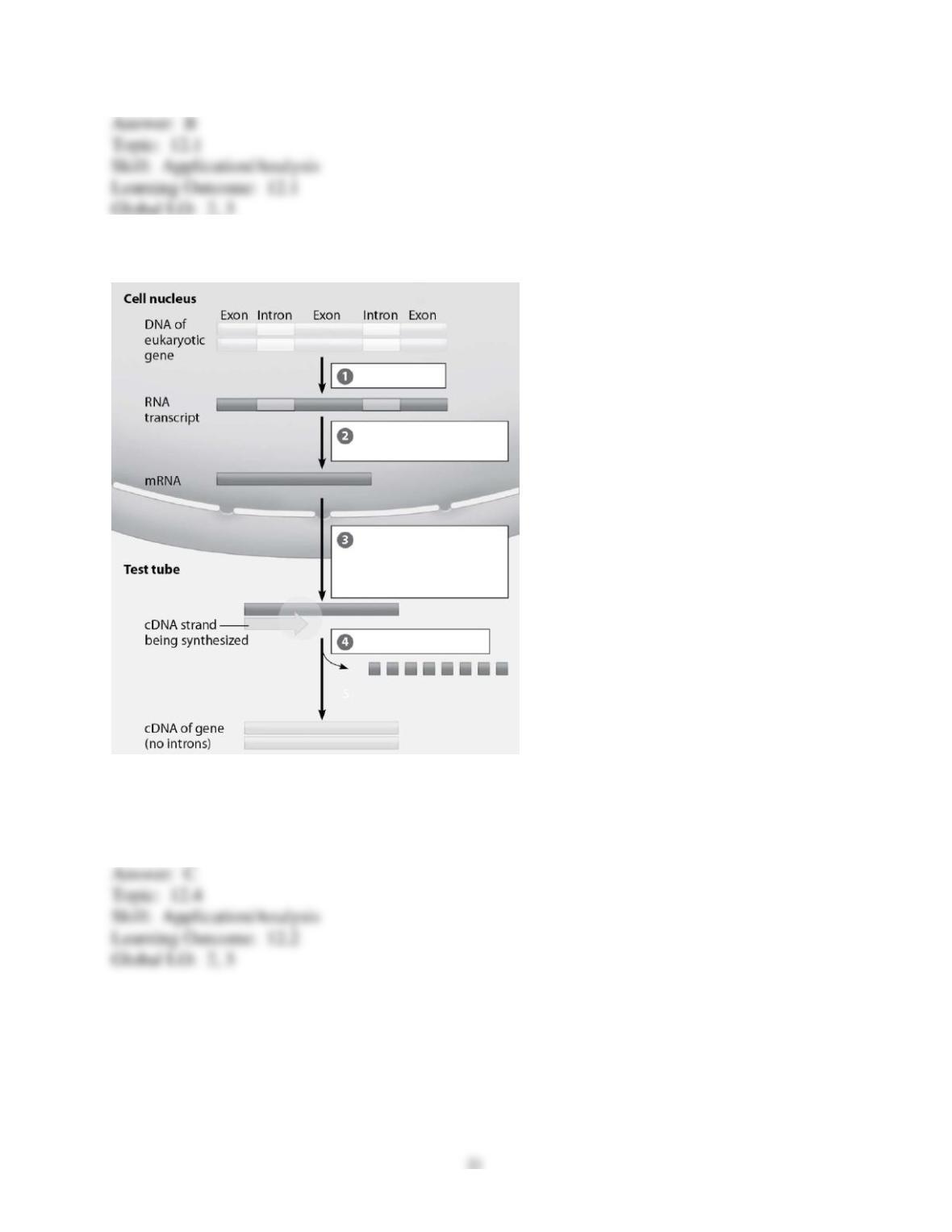
65) You prepare a genomic library and use this as a source to produce recombinant proteins in E.
coli. However, after producing the proteins you find that they are all too long or too short. In
fact, you don't find a single protein that is the correct size. What is the best explanation for this
result?
A) Since a genomic library was used as the source of the genes, the introns were included during
transcription and translation.
B) Since bacteria were used to produce the proteins, sugar groups were not properly added on to
the proteins that were being made.
C) Since a genomic library was used as the source, the recombinant bacterial plasmid DNA was
likely incorporated into the proteins being made.
D) Since bacteria were used to produce the proteins, the native bacterial plasmid DNA was likely
incorporated into the proteins being made.
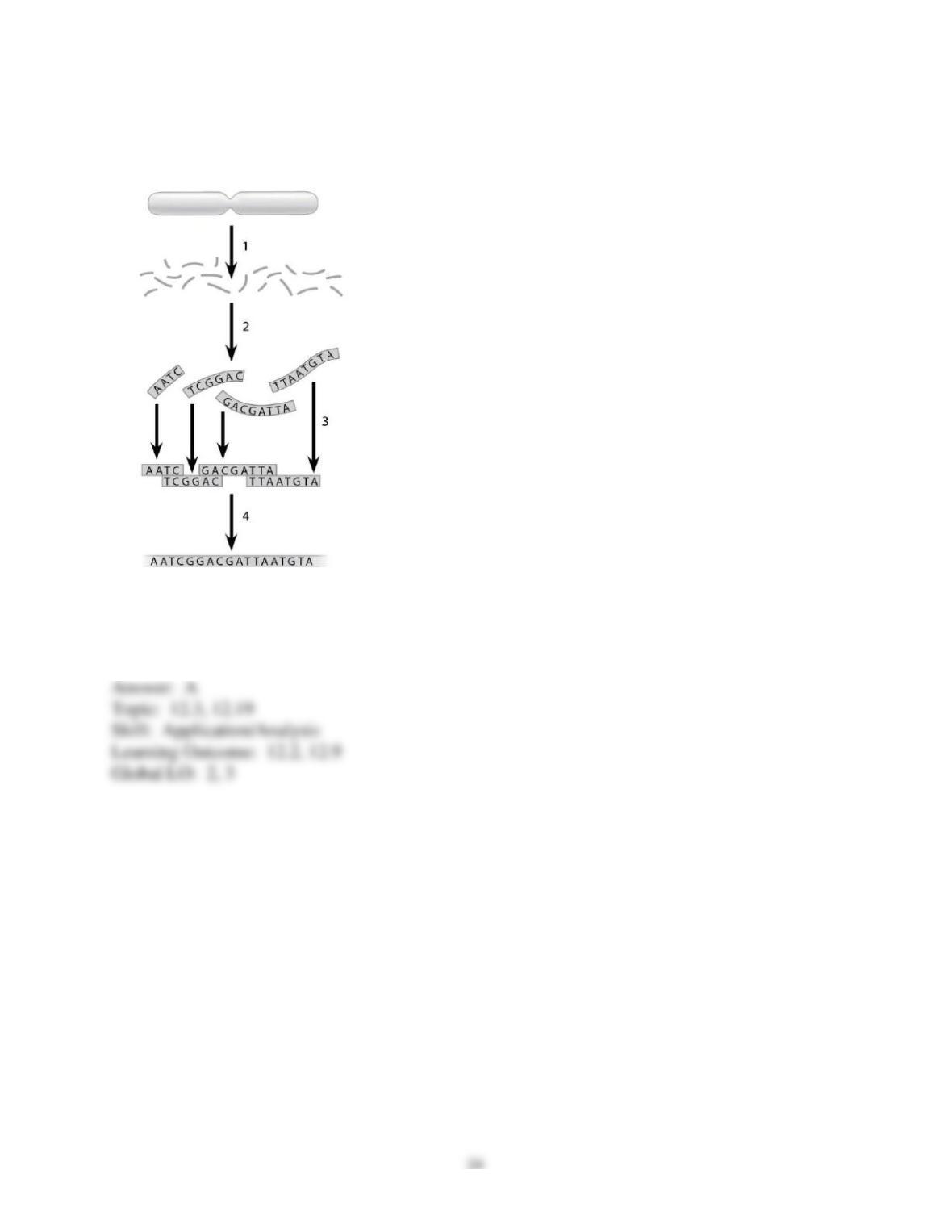
66) You are trying to produce a cDNA library starting from mRNA in skeletal muscle cells. You
add the following components to a tube and let the reaction proceed: skeletal muscle mRNA, free
nucleotides, and DNA polymerase. After inspecting the products of the reaction, you do not find
any cDNA molecules at all. What is the most likely explanation for this result?
A) You used mRNA instead of genomic DNA as the source material.
B) You used DNA polymerase instead of reverse transcriptase.
C) cDNA libraries can only be made from skin cell mRNA molecules.
D) You forgot to add a radiolabeled nucleic acid probe to the tube.
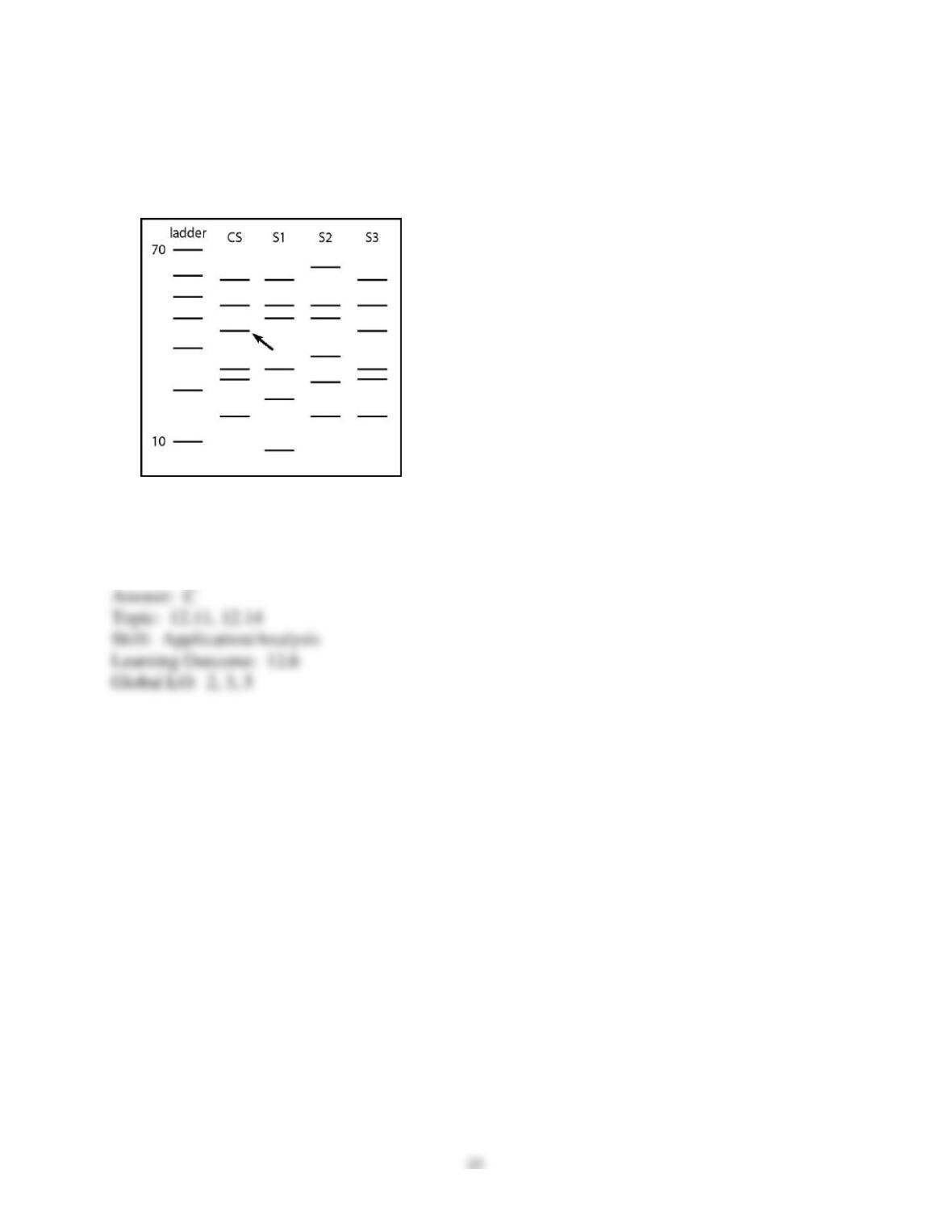
67) Gel electrophoresis is normally set up with the negative electrode at the top of the gel and the